Комбинирование задач связывания сущностей и извлечения отношений с использованием объединённой нейросетевой модели
Aннотация
В данной работе мы описываем методы обучения нейросетевых моделей для извлечения фармацевтически значимых сущностей из текстов на естественном языке с дальнейшим преобразованием их в формализованный вид тезаурусов и специализированных словарей, а также установления связей между ними. Задача извлечения соответствующей фармацевтической информации из интернет-текстов востребована органами фармаконадзора для мониторинга последствий и условий приема лекарственных средств. Анализ текстов из Интернета осложняется наличием неформальной речи и искаженной терминологии. Следовательно, анализ требует не только извлечения фармакологически значимой информации, но и приведения ее к стандартизированной форме. Целью данной работы является получение единой модели нейронной сети, которая решает все три задачи - распознавание сущностей, извлечение отношений и разрешения неоднозначности сущностей - чтобы избежать последовательной обработки одного текста независимыми моделями. Мы рассматриваем подходы, основанные на генеративных нейронных сетях, которые генерируют последовательности слов в соответствии с заданным входным текстом, и экстрактивных нейронных сетях, которые выбирают и классифицируют слова и последовательности в исходном тексте. Результаты сравнения показали преимущество экстрактивного подхода перед генеративным в рассматриваемом наборе задач. Модели этого подхода превосходят генеративную модель на 5% (f1-микро=85,9) в задаче извлечения фармацевтических объектов, на 10% (f1-микро=72,8) в задаче извлечения отношений и на 4% (f1-микро=64,5) в задаче разрешения неоднозначности. Также была получена совместная экстрактивная модель для трех задач с точностью f1-micro: 83,4, 68,2, 57,4 для каждой из задач.
Ключевые слова: Обработка естественного языка, Большие языковые модели, Фармация, Распознавание сущностей, Нормализация медицинских концептов, Разрешение неоднозначностей
К сожалению, текст статьи доступен только на Английском
INTRODUCTION
The Internet is filled with textual information that people exchange. Developing methods for extracting meaningful information from the total volume is an urgent challenge for natural language processing specialists. This work is devoted to the development of neural network models that solve the problems of extracting significant text fragments (entity extraction, ER), linking selected entities with each other (relation extraction, RE), linking selected entities with terms from the thesaurus (entity disambiguation, ED). This study was conducted in the field of extracting pharmaceutically relevant information from online drug reviews. The methods being developed may be useful, for example, for pharmacovigilance purposes. Automatic analysis of drug reviews allows one to monitor drugs that have already passed clinical trials and been released to the market. The purpose of such an analysis is to identify features of drugs that were not identified in clinical trials. For example, unwanted side reactions or unwanted effects that occur when different drugs interact. Automatic analysis of online reviews is complicated by the fact that texts from the Internet usually contain distorted terminology, errors, typos, etc.
In previous works, we proposed methods for extracting entities based on classical methods (Sboev et al., 2022a), a model for identifying relationships between entities (Sboev et al., 2023), and also tested a method for bringing medical entities to thesaurus terms based on the ranking method (Sboev et al., 2022b). The best results are obtained when using language models based on transformer architecture as encoders, which requires significant computing resources. Therefore, using these three separate models for sequential text processing is quite resource-intensive. In this regard, the goal of this work is to develop an end-to-end trained model that jointly solves the problem of extracting entities and bringing them to a standardized form according to thesauri, as well as establishing the relationships between them. Among pharmaceutically significant entities, we primarily consider the names of the drugs, the symptoms and diseases for which they were taken, and the adverse reactions they caused.
In our research we use large language models (LLM) (Devlin et al., 2018, Liu et al., 2019, Raffel et al., 2020). They are neural networks with a large number of parameters, usually pre-trained on a large number of texts using unsupervised techniques. These models are very popular nowadays because they have a high generalizing ability and are able to solve a wide range of problems by learning from fewer examples than models trained from scratch. However, such models are usually not very effective for tasks in specific domains (such as medicine), but can be used as a foundation that can be incorporated into other neural networks or fine-tuned for a particular task.
As part of the study, we compare extractive and generative approaches. We consider extractive methods to be models based on encoder LLM (which transforms the input text into a real-valued vector space) and the classification of words and fragments of the source text, in which case the information they contain is highlighted. These are the most common and widely used methods for extracting entities, relationships and facts from text. The generative approach is based on the use of generative language models trained on a large array of data and are capable of generating coherent text in response to an input prompt. Those models are usually bigger then encoder LLMs and are trained on bigger datasets, so they are able to solve simple task learning by just a few examples. But for a more complex task in specific domains they still should be trained to produce structured data extracted from the original analyzed text. The following is an analysis of the main works on this topic that use both approaches.
RELATED WORKS
Entity extraction methods. The task of extracting useful entities is a classic natural language processing task for which many methods have been developed. There are two common approaches to entity extraction: token classification (Liu et al., 2022) and span-based approach (Eberts, Ulges, 2020). When solving this task in texts of some specific domains, the choice of encoding method for input text tokens plays an important role. Currently, transformer language models encoders are leading in this area. General-purpose models like BERT (Devlin et al., 2018), RoBERTa (Liu et al., 2019), XLM-RoBERTa and others perform well on a wide variety of texts. However, when processing texts with a medical focus, variants of these models are required that are additionally trained on the corresponding corpora. For example, in one work (Lee et al., 2019) BioBERT was used, a version of the BERT model trained on articles from PubMed. For the task of analyzing Russian-language texts containing pharmaceutical information, there are two encoder models, pre-trained on corpora of reviews of medical products XLM-Roberta-large-sag (Sboev et al., 2022a) and RuDR-BERT (Tutubalina et al., 2021).
Relation extraction methods. The task of relationship extraction is usually reduced in the literature to classifying pairs of entities. Having two extracted entities, the neural network generates vector representations for each entity, after which a vector representation of the pair is formed using a combination of entity vectors (Sahu, Ashish, 2018; Sboev et al., 2022a), as well as additional features (Zhou et al., 2021). After this, the concatenated vector is fed into the classification layer, whose task is to predict the presence of a relationship and its type. Currently, the direction of research has shifted in favor of models that simultaneously solve the problem of extracting entities and establishing connections between them (Eberts, Ulges, 2020). In this case, for both tasks one needs to build context-sensitive vector representations, for which heavy language models are usually used, and there is no point in running the text through them twice.
Entity disambiguation methods. The task of entity disambiguation in the field of pharmacy is to establish a link between terms from specialised thesauri (e.g. UMLS, MedDRA, SNOMED-CT) and specific text fragments (usually in an informal form) containing references to pharmaceutically relevant entities. There are two groups of methods. Classification methods usually have a block (feature extractor) that converts text into a vector representation (embedding). This embedding is then passed through the Softmax layer, resulting in a probability distribution vector over the thesaurus terms (Lin et al., 2019; Pattisapu, 2020). Ranking-based methods use models that are trained to determine how semantically close a mention and a term are to each other. They do this by mapping the mention and the term into a multidimensional space Rn. Then the similarity (for example, cosine) is calculated for these vectors to determine which term the mention most closely matches (Sboev et al., 2022b; Mondal et al, 2019; Yuan et al, 2022; Sakhovskiy et al., 2023). Like the relation extraction task, the entity disambiguation task is also sometimes combined with the entity extraction stage. One of the first such works (Broscheit, 2020) proposes a straightforward end-to-end method that simultaneously searches for mentions and links them. Another work (De Cao et al., 2020) proposed a generative approach to collaborative mention detection and resolution.
Generative models. After BERT-like encoder models gained popularity, generative models began to appear that included a decoder in their architecture. In order for these models to learn to generate coherent, logical texts, they are made to have a very large number of parameters and are trained on very large samples of texts. This allows them to successfully solve problems of summarizing texts and answering questions, but they are also used to extract data from texts. For example, you can solve the problem of entity extraction by training the model to generate a list of entities that are mentioned in it for each input text (Kondragunta et al, 2023). Early models of this type, which fit well on 1 GPU and can be easily retrained for their target task, include models of the T5 and GPT families. Today, huge models like ChatGPT, LLama, mistral are extremely popular, but their additional training requires enormous resources. Therefore, in order to adapt them to specific tasks, one needs to use the so-called in-context learning. In this work, we decided to limit ourselves to the T5 model, which has shown its effectiveness on a wide range of tasks (Raffel et al, 2020) and can be fine-tuned.
DATA
Experiments were carried out on the Russian-language corpus of online drug reviews RDRS (Sboev et al, 2022a). The extended version of this corpus contains 3821 reviews, in which the annotators marked mentions of medical entities. The annotators were certified pharmacists, as well as students with pharmaceutical education. Statistics on the frequency of use of different types are shown in Table 1. To standardize the information extracted from the reviews, the MedDRA thesaurus was used – the Medical Dictionary for Regulatory Activities terminology is the international medical terminology developed under the auspices of the International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (ICH). We used PT level terms from it to standardize the mention of adverse reactions (ADR) and symptoms (Indication).
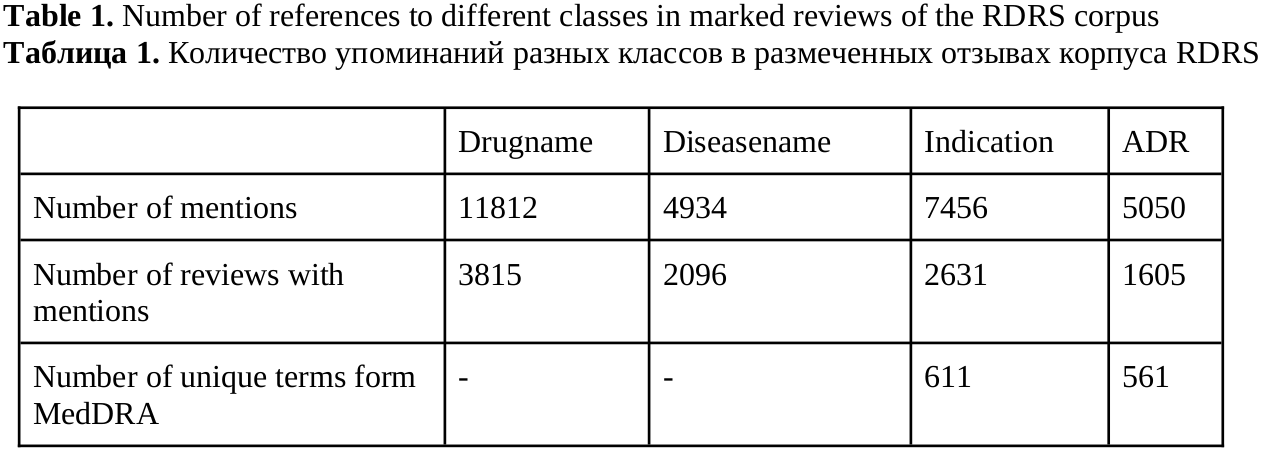
Most reviews are written about one drug (the main one). In some of the reviews, the authors mention several additional drugs used for different diseases and causing different adverse reactions at different times. In this regard, the entities are assigned either to the main context or to other, additional ones. If two entities belong to the same context, then they are related. We consider relations of the following types: Drugname–ADR (12717 relations in the corpus), Drugname–Diseasename (14147), Diseasename–Indication (9210).
METHODS
We compare approach methods for extracting and normalizing entities from review texts. The pipeline approach is the following: the method uses two neural network models, one for identifying entities from texts and establishing relations between them, the other for linking the selected entities with a thesaurus. The joint approach is based on the same models, but we combined them into a single model that learns all tasks at once. The generative method is based on a generative model with T5 architecture.
Pipeline. This method consists of sequential application of two models. The first one is a model based on the SpERT (Eberts, Ulges, 2020) architecture. We previously used it to identify pharmaceutically significant entities and the relationships between them (Sboev et al., 2023). The xlm-roberta-large-sag (Sboev et al, 2022a) model, pre-trained on a large corpus of unlabeled medical reviews, was used as an encoder.
The input text is divided into a sequence of tokens (words or parts of words), after which each token is encoded by the encoder language model and turned into a real-valued vector. Next, a set of possible spans is formed: sequences of tokens ranging in size from 1 to the maximum allowed. For each span, a vector sij is formed by concatenating maxpooling over the tokens included in the span and embedding the length of the span.
sij = maxpool(ti, ti+1, ..., tj) ⊕ Embedding(i – j), here t are token embeddings, i ≤ j; j – i ≤ max span length.
The span vectors, together with the vector of the special token [CLS] which is added to the beginning of the text and is a vector representation of the entire text, enter the fully connected linear layer (Dense) with the number of output neurons corresponding to the number of entity types:
Cent{M, c+1} = Softmax(Dense(sij ⊕ t[CLS])), where Cent{M, c+1} is the correspondence matrix between span and entity class, c + 1 is the number of possible classes, including the “non-entity” class, M is the maximum number of spans. For the entity extraction task, the following loss function is calculated:, where Yent is a matrix of one-hot vectors in which the desired class is marked by one, and M entities include both positive examples (entities identified by annotators) and negative ones (randomly selected spans from the text, which must correspond to the zero class “non-entity”), and C is, respectively, a matrix where the original one-hot vectors correspond to the normalized confidence of the neural network in the classes for each span. Pairs of span vectors are then concatenated with each other with a result of maxpooling token vectors between them to classify the relationship for that pair of spans.
vreli j i' j' = sij ⊕ maxpool(tj, tj+1, ..., ti'-1) ⊕ si' j'.
For the relationship task, the loss function is also calculated:
Lrel = CrossEntropy(Yrel{N, r+1}, Crel{N, r+1}), where Yrel and Crel are, respectively, the reference one-hot vectors and the confidence of the neural network for the relationship classes of N pairs of spans. Span pairs, like entities, include positive and negative examples. In our case, only two classes of relations are considered – presence and absence of a relation.
Once the entities have been extracted, ADR and Indication entities are selected from them. Next, using the ranking method, suitable terms from MedDRA are selected for them. The ranking method was also trained according to prior work (Sboev et al., 2022b). This method encodes PTs from MedDRA using a language model tuned to produce vectors that have high cosine proximity for similar terms and low cosine proximity for different terms. After this, another language model, ruBERT (Zmitrovich et al., 2023) learns to produce vectors for mentions from the text that will also be close to the terms that the labelers assigned them, and far from all other terms. The loss function to be minimized is as follows:
Lnorm = CrossEntropy(Ynorm, Softmax(cos(Vent, VPT))), where the cross-entropy is calculated for the matrix of one-hot vectors that, for each mention from the batch, point to its corresponding PT from MedDRA, and the cosine proximity between the vectors of these mentions and the PT vectors normalized by Softmax. To encode PT MedDRA, we used the ruBERT model fine-tuned on translated mention-PT pairs from English corpora: CADEC, TAC 2017, SMM4H 2017, MedMention + PsyTAR.
Joint model. The combination of the Spert model and the ranking method was carried out as follows. After possible spans of entities have been compiled, in the section of the neural network where maxpooling is calculated to obtain the span vector, a branch is made in which meanpooling is calculated instead. The mean-pooled vector is passed through two layers with ReLU activation and the third linear layer which gives us resulting vectors to calculate similarity between mention and encoded PT terms from MedDRA. The loss functions for each individual task are calculated in the same way as in the pipeline approach, but the final function contains coefficients for balancing. This is needed due to the fact that in the SpERT model training mode, there are much fewer examples for training in a batch than when training the model separately for normalization. Thus Ljoint = Lent + Lrel + αLnorm. The best results were obtained with α = 100.
Generative LLM. Generative large language models are capable of predicting the most likely sequence of tokens in response to an input prompt. The model with the T5 (Raffel et al., 2020) transformer encoder-decoder architecture (large version) contains 24 transformer blocks in the encoder and decoder. The Russian version of the model (Zmitrovich et al., 2023) was trained on a large corpus of Russian texts for the task of predicting masked spans. We took this model as a basis and trained it for the task of extracting related entities and their corresponding terms. Given the text of reviews as input, the model had to generate an HTML-like result: <Indication MedDRA=Insomnia context=main>I can’t sleep</Indication>, <ADR MedDRA=Sore throat context=other>throat hurts after taking it</ADR>.
EXPERIMENTS AND RESULTS
During the experiments, each method was required to provide for every input text the following: a set of entities and their classes (Drugname, Diseasename, ADR, Indication), a set of relations for entities Drugname-ADR, Drugname-Diseasename, Diseasename-Indication, for each entity of type ADR or Indication must be consistent with the term MedDRA.
To evaluate the quality of entity extraction, the following metrics are used: Precision, Recall, F1. They are calculated taking into account the boundaries of true and predicted entities and their classes. The same metrics are used to evaluate the quality of relationship extraction. But pairs of entities with classes were taken into account. Triples (entity boundaries, entity class, thesaurus term) are used to calculate f1 for the ambiguity resolution problem.
For each problem, metrics are calculated with micro (calculated paying attention to class imbalance) and macro (mean of scores across the classes) averaging. Experiments were carried out using cross-validation using 5 folds. The resulting metrics provided in Table 2.
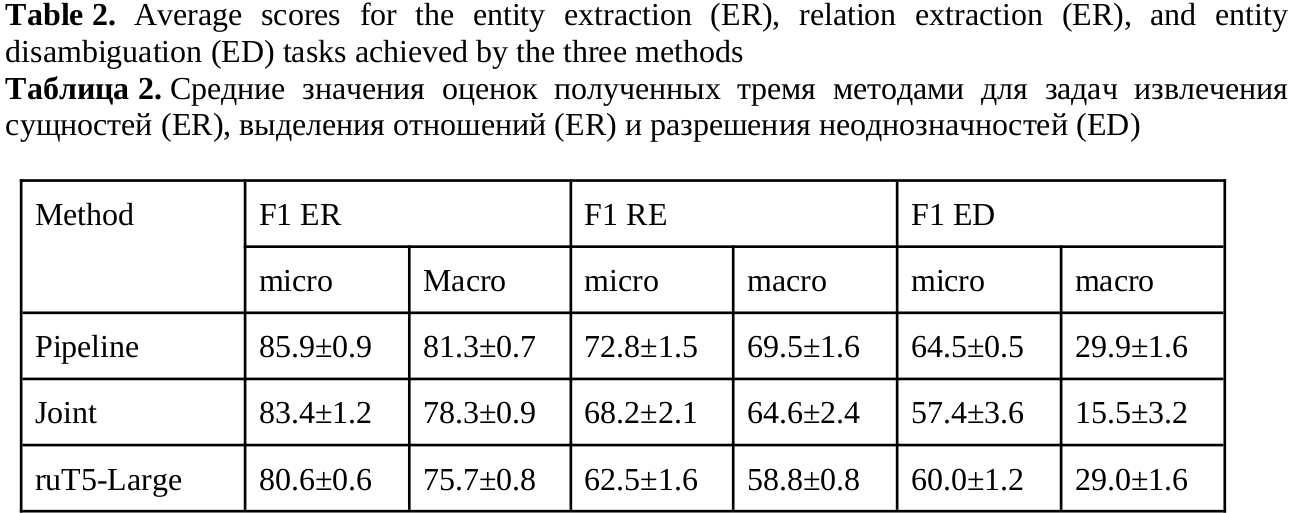
Both variants of the end-to-end model (joint and ruT5 methods) showed a decrease in accuracy compared to the sequential approach. A particularly strong drop in accuracy is observed in f1-macro for the joint model ambiguity resolution task, this suggests that it is trained only on PTs, for which there are many examples in the texts. It is worth noting that in the sequential version there is a big difference in the training procedures of the original models. The ambiguity resolution model is trained on minibatches consisting of 32 mention-term pairs. And in the combined version, each minibatch contains only 2 texts, which may not contain entities like ADR and Indication at all. This difference was slightly compensated by selecting the balancing coefficient for the loss functions. The difference in encoder models for entities also has an effect: in the sequential approach, mentions from texts are encoded using ruBERT, and PT using fine-tuned ruBERT, while in the combined model, the xlm-roberta-large-sag model is left for encoding mentions for better accuracy of entity extraction and relationships. The ruT5 model lags behind the sequential approach in all evaluations except f1-macro ambiguity resolution. This is where the great generalizing ability of this model comes into play.
DISCUSSION
The main reason for drawback in all scores is the class imbalance in the corpus. For entity recognition we provide a table with detailed scores (Table 3). Looking at Table 1, the ADR class is difficult to predict because of its low representativeness, but it is also the most difficult class because there are more ways to say "headache" or "abdominal pain" than to distort the name "aspirin".
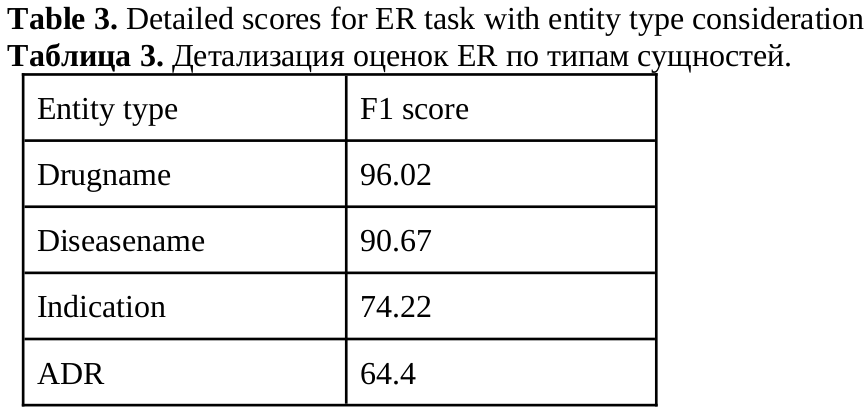
For entity recognition the class imbalance is not very serious and it’s shifted to positive examples. Every document has positive relations between entities, but there are 1328 documents in which some entities are not related to each other.
For entity disambiguation the PTs imbalance is very strong. Of the 860 different PTs in the corpus, 20% appear only once in the corpus, so they can only be present in training or test, 55% appear less than 10 times, and only 25% of the PTs appear more than 10 times. None of the models can successfully recognize PTs of the first group. For PTs that appear 2-10 times in corpus have critically low average f1 score 1.5%. And only 25% of the most common PTs can be correctly classified, with an average score of 40%. That explains the difference between micro and macro scores and low scores in general.
Another problem that lower the scores for relation extraction and entity disambiguation is the fact that results are applied to the result of entity recognition so there is a combined error for these tasks. The application of this methods to the true entities without entity recognition step were analyzed in our previous works (Sboev et al., 2023, Sboev et al., 2022b) which can be considered as state-of-the-art for this corpus.
We also did a small test on 1 of the folds test sets, for chatGPT-3.5. But out of the box the performance was very bad, the entity recognition score for ADR and Indication was only 29%. It usually extracts only the first occurrence of mentions without extracting their repetitions, and even in those the boundaries were sometimes different.
Here is an example of predictions for the text.
Original text:
Чем выкидывать кучу денег на лечения, я предпочитаю проводить профилактические меры. Пользоваться этой мазью я начале еще в училище. Так как училась я в медицинском училище учителя, особенно в зимнее время, чтобы студенты не пропускали занятия по болезни побуждали нас утречком перед занятием мазать в носу Оксолинкой. Мазь действительно помогает. Хотя есть в ней и недостаток. Сразу после нанесения в носу немного печет и чувство заложенности. Однако через время это проходит. Слизистая оболочка не пересыхает, благодаря мази она всегда увлажненная. И к тому же защищает организм от инфекций. Данную мазь можно применять даже детям. Если вы тоже за профилактику заболеваний, то рекомендую воспользоваться данной противовирусной мазью.
English translation:
Rather than throwing away a lot of money for treatment, I prefer to take preventive measures. I started using this ointment back in college. Since I studied at the medical school, teachers, especially in winter, so that students would not miss classes due to illness, encouraged us to smear Oxolink in our nose in the morning before class. The ointment really helps. Although there is a drawback in it. Immediately after application, the nose bakes a little and a feeling of stuffiness. However, it passes after a while. The mucous membrane does not dry out, thanks to the ointment it is always moistened. And besides, it protects the body from infections. This ointment can be used even for children. If you are also for the prevention of diseases, then I recommend using this antiviral ointment.
True entities:
Оксолиновая мазь (Drugname), профилактические (Indication, PT=Профилактика), Оксолинкой (Drugname), в носу немного печет (ADR PT=Дискомфорт в носу), чувство заложенности (ADR PT=Заложенность носа), профилактику (Indication PT=Профилактика)
Entities extracted by joint method:
Оксолиновая (Drugname), Оксолиновая мазь (Drugname), профилактические (Indication PT=Профилактика), Оксолинкой (Drugname), чувство заложенности (ADR PT=Заложенность носа), заложенности (ADR PT=Заложенность носа), профилактику (Indication PT=Профилактика)
Entities extracted by generative method:
Оксолиновая мазь (Drugname), профилактические (Indication PT=Профилактика), Оксолинкой (Drugname), в носу немного печет (ADR PT=Боль в носу), чувство заложенности (ADR PT=Заложенность носа), профилактику (Indication PT=Профилактика)
CONCLUSIONS
A comparison is made of an extractive approach based on models whose architecture is designed specifically for extracting data from the input text and a generative approach, in which a general-purpose generative model is additionally trained for the task of generating structured text containing the required information from the source text. The results show that the generative model is much easier to use, but loses in accuracy in such a specific task. The extractive sequential approach surpasses it in accuracy by 5% in the entity extraction task, 10% in the relationship extraction task, and 4% in the ambiguity resolution task and allows us to obtain the following accuracies for solving three problems: 85.9, 72.8, 64.5. The combined extractive model has lower accuracy than the sequential approach: 83.4, 68.2, 57.4.
The work was carried out within the state assignment of NRC "Kurchatov Institute" using computing resources of the federal collective usage center Complex for Simulation and Data Processing for Mega-science Facilities at NRC “Kurchatov Institute”, http://ckp.nrcki.ru/.



















Список литературы
Список использованной литературы появится позже.