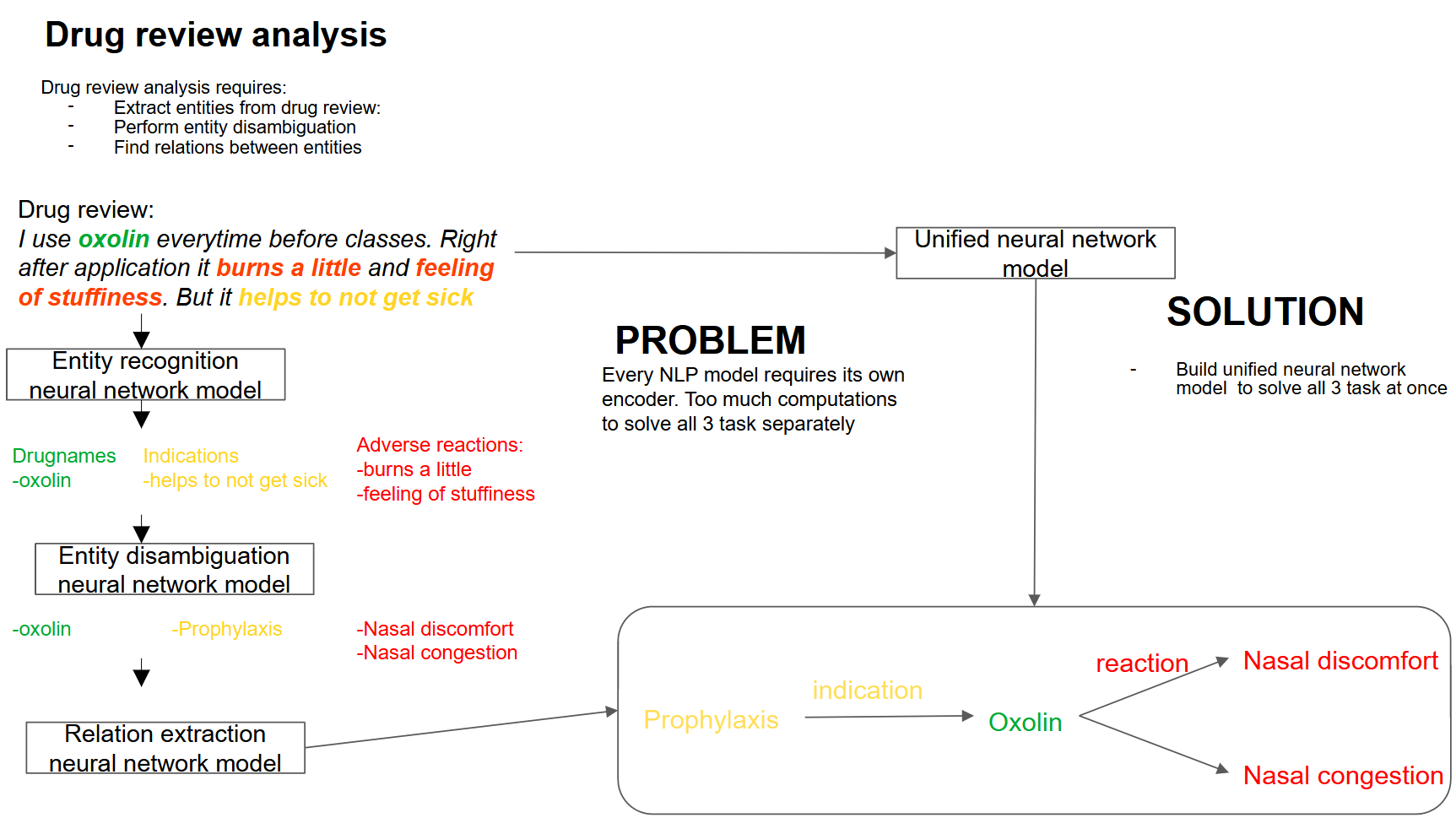
Combining the tasks of entity linking and relation extraction using a unified neural network model
In this paper we describe methods for training neural network models for extracting pharmacologically significant entities from natural language texts with their further transformation into a formalized form of thesauruses and specialized dictionaries, as well as establishing relations between them. The task of extracting relevant pharmaceutical information from Internet texts is in demand by pharmacovigilance to monitor the effects and conditions of taking medicines. The analysis of texts from the Internet is complicated by the presence of informal speech and distorted terminology. Therefore, the analysis requires not only extracting pharmacologically relevant information, but also bringing it to a standardized form. The purpose of this work is to obtain an end-to-end neural network model that solves all three tasks – entity recognition, relation extraction, and entity disambiguation – in order to avoid sequential processing of one text by independent models. We consider approaches based on generative neural networks that create sequences of words according to a given input text and extractive ones that select and classify words and sequences within the source text. The results of the comparison showed the advantage of the extractive approach over the generative one on the considered set of tasks. The models of this approach outperform the generative model by 5% (f1-micro=85.9) in the task of extracting pharmaceutical entities, by 10% (f1-micro=72.8) in the task of extracting relations and by 4% (f1-micro=64.5) in the entity disambiguation. A joint extractive model was also obtained for three tasks with f1-micro accuracy: 83.4, 68.2, 57.4 for each of the tasks.
Figures
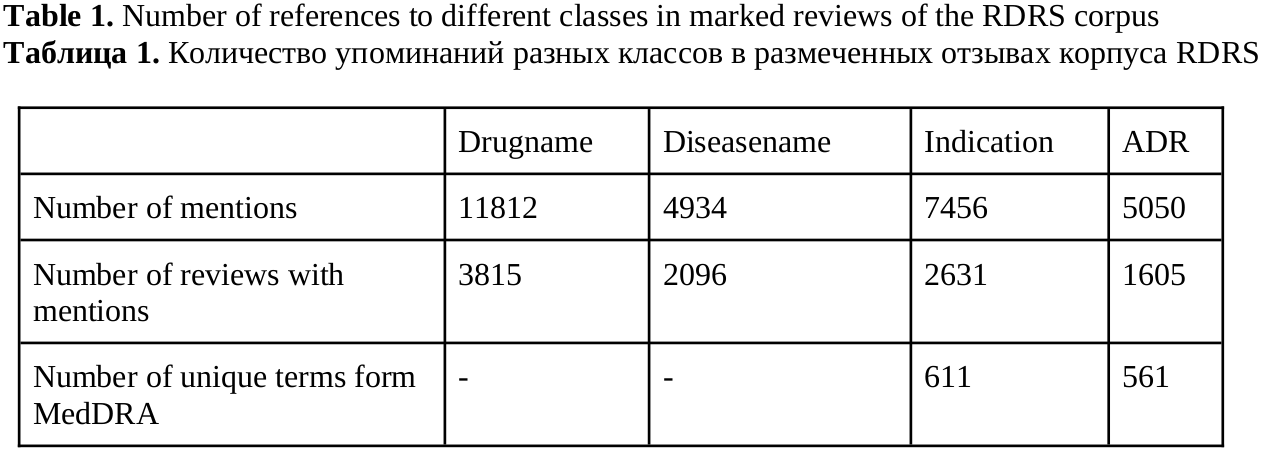
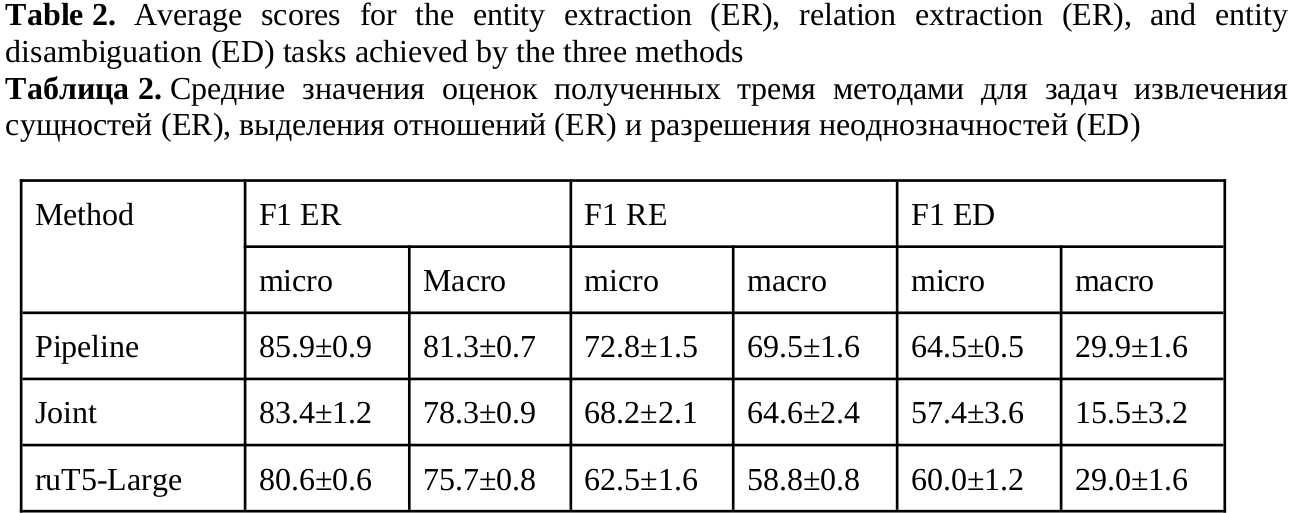
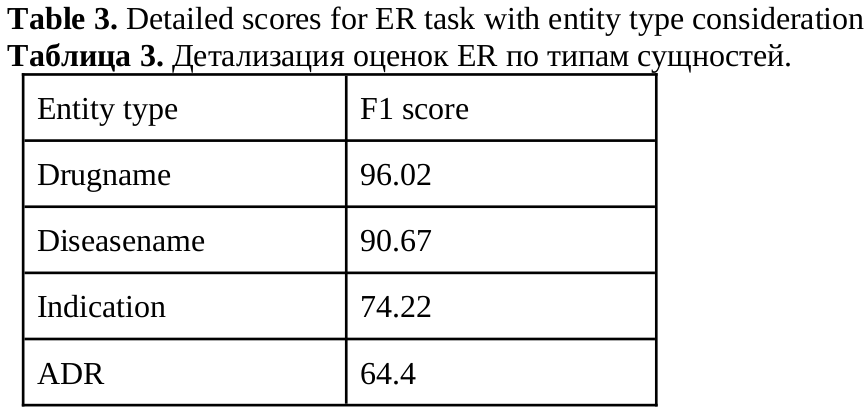
Sboev, A. G., Gryaznov, A. V. (2024). Combining the tasks of entity linking and relation extraction using a unified neural network model, Research Result. Theoretical and Applied Linguistics, 10 (4), 94-105.


















While nobody left any comments to this publication.
You can be first.
Broscheit, S. (2020). Investigating entity knowledge in BERT with simple neural end-to-end entity linking, arXiv preprint, arXiv:2003.05473. https://doi.org/10.18653/v1/K19-1063(In English)
De Cao N, Izacard G, Riedel S, Petroni F. (2020). Autoregressive Entity Retrieval, ICLR 2021 – 9th International Conference on Learning Representations, Vienna, Austria. (In English)
Devlin, J., Chang, M.-W., Lee, K. and Toutanova, K. (2019). BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding, arXiv:1810.04805v2. DOI: 10.48550/arXiv.1810.04805 (In English)
Eberts, M. and Ulges, A. (2020). Span-based joint entity and relation extraction with transformer pre-training, ECAI 2020, 325, 2006–2013. DOI: 10.3233/FAIA200321 (In English)
Kondragunta, M., Perez-de-Viñaspre, O. and Oronoz, M. (2023). Improving and simplifying template-based named entity recognition, Proceedings of the 17th Conference of the European Chapter of the Association for Computational Linguistics: Student Research Workshop, Dubrovnik, Croatia, 79–86. DOI: 10.18653/v1/2023.eacl-srw.8 (In English)
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H. and Kang, J. (2019). BioBERT: A pre-trained biomedical language representation model for biomedical text mining, Bioinformatics, 36, 4, 1234–1240. DOI: 10.1093/bioinformatics/btz682 (In English)
Lin, C., Lou, Y. S., Tsai, D. J., Lee, C. C., Hsu, C. J., Wu, D. C., Wang, M. C. and Fang, W. H. (2019). Projection word embedding model with hybrid sampling training for classifying ICD-10-CM codes: Longitudinal observational study, JMIR medical informatics, 7 (3), e14499. DOI: 10.2196/14499 (In English)
Liu, Y, Ott, M, Goyal, N, Du, J, Joshi, M, Chen, D, Levy, O, Lewis, M, Zettlemoyer, L, Stoyanov, V. (2019). Roberta: A robustly optimized BERT pretraining approach, arXiv preprint. https://doi.org/10.48550/arXiv.1907.11692(In English)
Liu, P., Guo, Y., Wang, F. and Li, G. (2022). Chinese named entity recognition: The state of the art, Neurocomputing, 473, 37–53. https://doi.org/10.1016/j.neucom.2021.10.101(In English)
Mondal, I., Purkayastha, S., Sarkar, S., Goyal, P., Pillai, J., Bhattacharyya, A. and Gattu, M. (2019). Medical entity linking using triplet network, Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, Minnesota, USA, 95–100. DOI: 10.18653/v1/W19-1912 (In English)
Pattisapu, N., Anand, V., Patil, S., Palshikar, G. and Varma, V. (2020). Distant supervision for medical concept normalization, Journal of biomedical informatics, 109, 103522. DOI: 10.1016/j.jbi.2020.103522 (In English)
Raffel, C., Shazeer, N., Roberts, A., Lee, K., Narang, S., Matena, M., Zhou, Y., Li, W. and Liu, PJ. (2020). Exploring the limits of transfer learning with a unified text-to-text transformer, Journal of machine learning research, 21 (14), 1–67. (In English)
Sahu, S. K. and Ashish, A. (2018). Drug-drug interaction extraction from biomedical texts using long short-term memory network, Journal of biomedical informatics, 86, 15–24. DOI: 10.1016/j.jbi.2018.08.005 (In English)
Sakhovskiy, A., Semenova, N., Kadurin, A. and Tutubalina, E. (2023). Graph-enriched biomedical entity representation transformer, Experimental IR Meets Multilinguality, Multimodality, and Interaction,CLEF 2023. Lecture Notes in Computer Science, 14163, Springer, Cham. DOI: 10.1007/978-3-031-42448-9_10 (In English)
Sboev, A., Sboeva, S., Moloshnikov, I., Gryaznov, A., Rybka, R., Naumov, A., Selivanov, A., Rylkov, G. and Ilyin, V. (2022a). Analysis of the full-size Russian corpus of internet drug reviews with complex NER labeling using deep learning neural networks and language models, Applied Sciences, 12.1, 491. DOI: 10.3390/app12010491 (In English)
Sboev, A., Rybka, R., Gryaznov, A., Moloshnikov, I., Sboeva, S., Rylkov, G. and Selivanov, A. (2022b). Adverse Drug Reaction Concept Normalization in Russian-Language Reviews of Internet Users, Big Data and Cognitive Computing, 6 (4), 145. DOI:/10.3390/bdcc6040145 (In English)
Sboev, A., Rybka, R., Selivanov, A., Moloshnikov, I., Gryaznov, A., Naumov, A., Sboeva, S., Rylkov, G. and Zakirova, S. (2023). Accuracy analysis of the end-to-end extraction of related named entities from Russian drug review texts by modern approaches validated on English Biomedical corpora, Mathematics, 11 (2), 354. DOI: 10.3390/math11020354 (In English)
Tutubalina, E., Alimova, I., Miftahutdinov, Z., Sakhovskiy, A., Malykh, V. and Nikolenko, S. (2021). The Russian Drug Reaction Corpus and neural models for drug reactions and effectiveness detection in user reviews, Bioinformatics, 37 (2), 243–249. DOI: 10.1093/bioinformatics/btaa675 (In English)
Yuan, Z., Zhao, Z., Sun, H., Li, J., Wang, F. and Yu, S. (2022). CODER: Knowledge-infused cross-lingual medical term embedding for term normalization, Journal of biomedical informatics, 126, 103983. DOI: 10.1016/j.jbi.2021.103983 (In English)
Zhou, W., Huang, K., Ma, T. and Huang, J. (2021). Document-level relation extraction with adaptive thresholding and localized context pooling, Proceedings of the AAAI conference on artificial intelligence, 35 (16), 14612-14620. DOI: 10.1609/aaai.v35i16.17717 (In English)
Zmitrovich, D., Abramov, A., Kalmykov, A., Tikhonova, M., Taktasheva, E., Astafurov, D., Baushenko, M., Snegirev, A.,
Kadulin, V., Markov, S. and Shavrina, T. (2023).
A family of pretrained transformer language models for Russian, arXiv preprint, arXiv:2309.10931. DOI: 10.48550/arXiv.2309.10931 (In English)